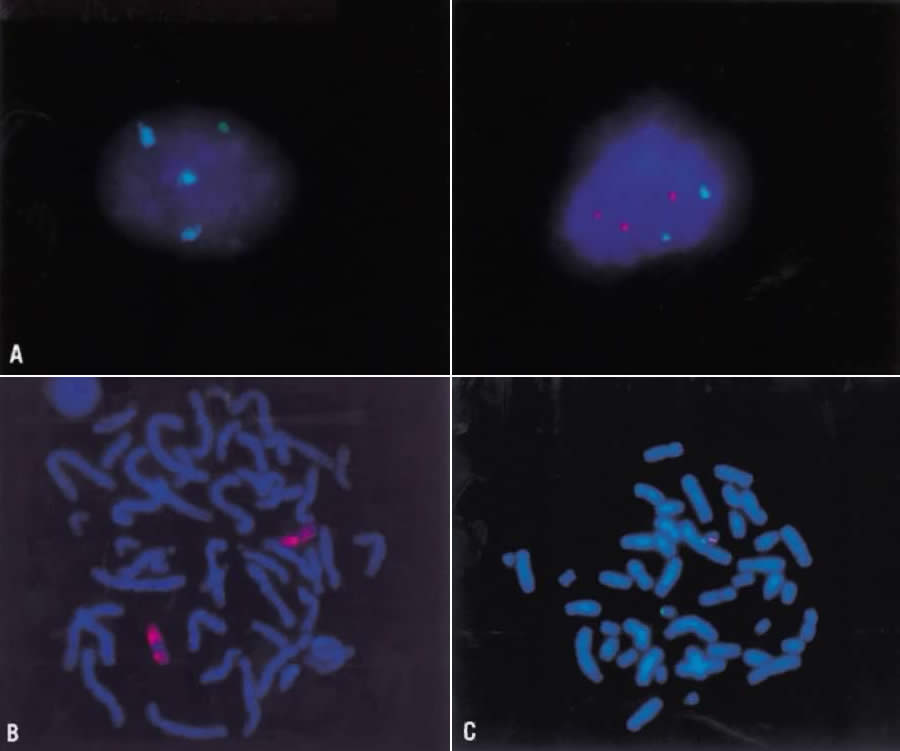
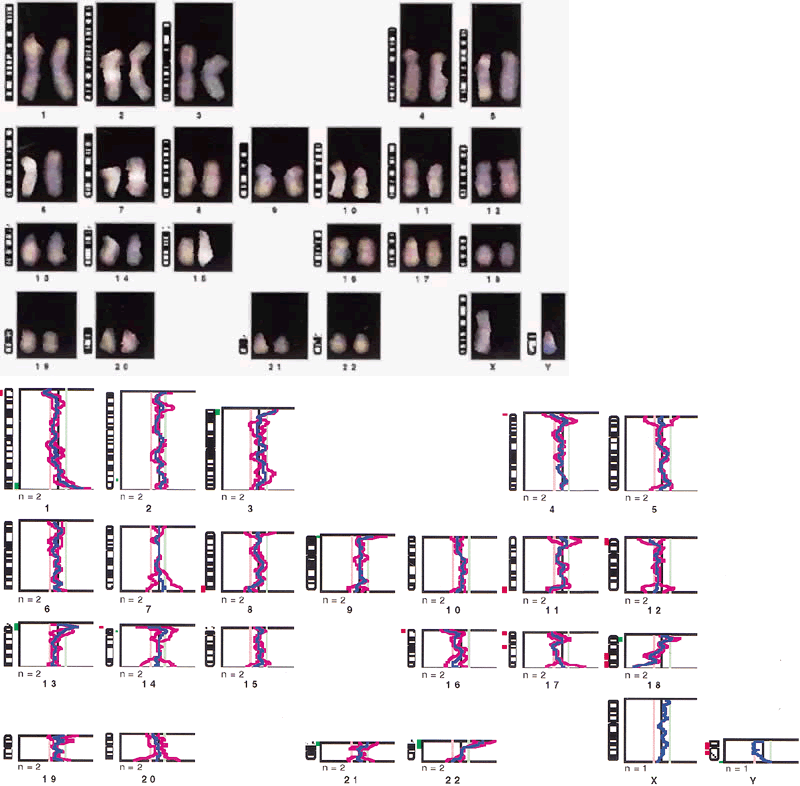
The normal human chromosome complement represents a balanced genetic constitution: the
correct chromosome number and the correct gene content. Deviations
from the normal number of chromosomes (i.e. numeric aberrations) or the normal morphologic pattern (i.e. structural rearrangements) represent gains or losses of genetic information
from the complement. Genetic imbalances generally disturb the normal
developmental processes of fertilization and embryogenesis and result
in implantation failure, spontaneous abortion, stillbirth, or a liveborn
infant with multiple congenital anomalies. It is possible for
a person to carry a chromosome rearrangement but show no clinical effects
because there is no accompanying genetic imbalance. Such an individual
is called a balanced carrier because the chromosome aberration constitutes a rearrangement of the genetic
material into a new chromosome pattern, with no gain or loss of
genes. Nevertheless, a balanced carrier is at reproductive risk to transmit
the chromosome rearrangement to offspring by gametes that are balanced
or unbalanced. A problem arises when prenatal diagnosis reveals
the presence of a de novo chromosome rearrangement, because it may not be possible to determine
whether or not such a rearrangement is accompanied by a microscopically
undetectable gain or loss of genetic material. Numeric Aberrations POLYPLOIDY. The two common forms of polyploidy in human tissues are triploidy, in
which each haploid set of 23 chromosomes is present three times (3N = 69) (Fig. 6), and tetraploidy, in which each haploid set is present four times (4N = 92) (Fig. 7). An increased incidence of triploidy and tetraploidy has been reported
when oral contraceptive use was stopped within 6 months before conception,18 but this finding has not been confirmed. One percent of all human conceptions
are estimated to be triploids. In 66% of the cases, triploidy
arises from simultaneous fertilization of an oocyte by two spermatozoa. In
the remaining cases, triploidy arises from unreduced oocytes or spermatozoa (i.e. during gametogenesis, the chromosome number is not reduced from 46 to 23).19 Tetraploidy usually arises as a postzygotic event through the failure
of cytokinesis (division of the cytoplasm) after chromosome duplication
and the incorporation ofdaughter chromosomes into the same cell. Other
less likely mechanisms include trispermy and fertilization of a diploid
ovum by two haploid spermatozoa or by a diploid spermatozoon. 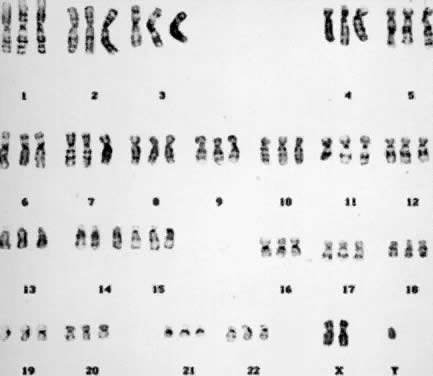 Fig. 6. Triploid (3N) karyotype 69,XXY (trypsin-Giemsa stain). Fig. 6. Triploid (3N) karyotype 69,XXY (trypsin-Giemsa stain).
|
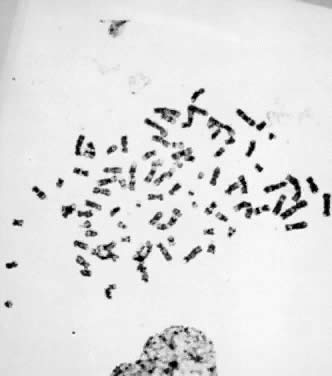 Fig. 7. Tetraploid (4N) karyotype 92,XXYY (trypsin-Giemsa stain). Fig. 7. Tetraploid (4N) karyotype 92,XXYY (trypsin-Giemsa stain).
|
A spectrum of clinical effects are associated with polyploidy. Based on
studies of preimplantation human embryos, most triploid embryos do not
continue to divide and to achieve the blastocyst stage. Triploid embryos
that implant frequently abort spontaneously in the first trimester; triploid
pregnancies constitute a major etiologic class within the
spontaneous abortion population because 15% to 20% of all chromosomally
abnormal spontaneous abortuses are triploid.20 A triploid complement has the potential of producing placental hydatidiform
degeneration and abnormal fetal development.21,22 Partial hydatidiform mole is found in 80% of triploids.23,24 The roles of the maternal and paternal genome in embryonic development
is illustrated in triploidy by comparing the effects of two sets of genes
from one parent and one set from the other. For example, when two
of the three sets in a triploid are derived from the maternal parent, fetal
development is enhanced to the detriment of the placenta. In contrast, when
two sets are derived from the paternal parent, placental
development is significantly favored over that of fetal development. A triploid chromosome syndrome has been defined, based on a series of developmental
anomalies observed in fetuses aborted electively after prenatal
diagnosis and in those rare cases of triploidy that complete pregnancy.22 These anomalies include severe intrauterine growth retardation (IUGR), hydrocephaly, holoprosencephaly, cortical and cerebellar hypoplasia, syndactyly, short
halluces, and rocker-bottom feet, and many other physical
defects. Chromosome analysis of triploid liveborn infants frequently
shows the presence of a normal cell line with 46 chromosomes, which
may be responsible for completion of pregnancy but not the prevention
of congenital malformations and severe developmental delay. Tetraploidy frequently is associated with spontaneous abortion in the first
trimester, although at least 10 cases of liveborn infants, with chromosome
mosaicism in five, have been reported.25 The latter were associated with low birth weight, microcephaly, microphthalmia
or anophthalmia, hypertelorism, low-set ears with dysplastic
cartilage, micrognathia, arachnodactyly, genital ambiguity, hypotonia, congenial
heart disease, and open neural tube defects. Death within 3 months
of birth was common, although some newborns with tetraploid mosaicism
survived longer. In the course of a chromosome analysis performed
on a viable fetus, it is not uncommon to observe complete tetraploidy
or diploid/tetraploid mosaicism in chorionic villi and amniotic fluid
cells. Almost all of these cases represent cultural artifacts or chromosome
changes limited to the sampled tissues and not the fetus proper. Fetal blood sampling by PUBS may be appropriate for fetuses whose ultrasound
evaluations demonstrate abnormal development. The decision to undertake
PUBS in all cases of tetraploidy does not have widespread agreement, nor
is it considered a standard of care, because PUBS is not without
obstetric risks and complications. Tetraploidy occasionally may occur in solid tumors.26 It does not appear to be specifically related to a particular tumor type, and
its relation to cancer is unknown. A series of mutations in genes
controlling cell cycle arrest after DNA damage were found to contribute
to tumor development by allowing the accumulation of chromosome
aberrations and gene mutations leading to cancer.27 These and other biologic processes that impair cytokines may be responsible
for the appearance of tetraploidy during carcinogenesis. Alternatively, because
polyploidy increases the total gene content, tetraploidy
may be one form of cellular response to the development of cancer (i.e. by increasing the total number of genes, a cell may have an advantage
over its diploid counterpart). ANEUPLOIDY. Aneuploidy, which is the gain or the loss of single, whole chromosomes, constitutes
the most important class of chromosome aberrations contributing
to human disease. The presence of only one member of a pair of
autosomal chromosomes (monosomy) invariably is lethal early in development. Monosomy for the X chromosome (45,X) is
the most common chromosome aberration in humans, occurring
in up to 5% of all conceptions and 15% to 25% of all first-trimester
spontaneous abortions with a chromosome aberration (Fig. 8).28 In 75% to 80% of the cases of monosomy X, nondisjunction of the X and
Y chromosomes during spermatogenesis is responsible. Other mechanisms
include nondisjunction during oogenesis or after fertilization, with the
latter resulting in chromosome mosaicism (e.g. 45,X/46,XX). The clinical features of 45,X are small stature; ovarian
dysgenesis, with concomitant amenorrhea and sterility; lymphedema, with
puffiness of the dorsum of the hands and feet; shield-like chest, frequently
with pectus excavatum; low posterior hairline; cubitus valgus; renal
anomalies, particularly horseshoe kidney; redundant skin of the
neck or webbed neck; and cardiac and aortic anomalies.28 In cases of chromosome mosaicism, the clinical effects of the monosomic
cell line may be mitigated by normal cells. 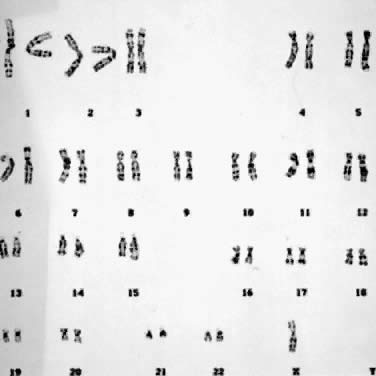 Fig. 8. Karyotype of 45,X. Gonadal dysgenesis (Turner) syndrome (trypsin-Giemsa
stain). Fig. 8. Karyotype of 45,X. Gonadal dysgenesis (Turner) syndrome (trypsin-Giemsa
stain).
|
The presence of an additional autosomal chromosome is referred to as trisomy, and it usually results from nondisjunction during gametogenesis. Premature
separation of chromosomes with random distribution may actually be
the underlying mechanism leading to aneuploid gametes. In trisomic conceptions, 1 of
the 22 autosomal chromosomes is present three rather
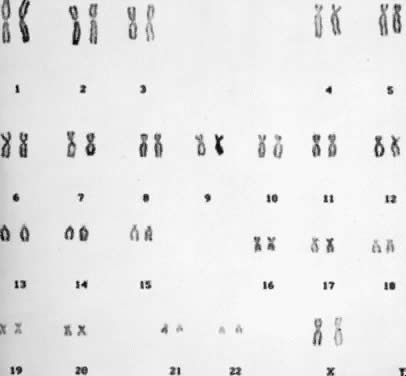
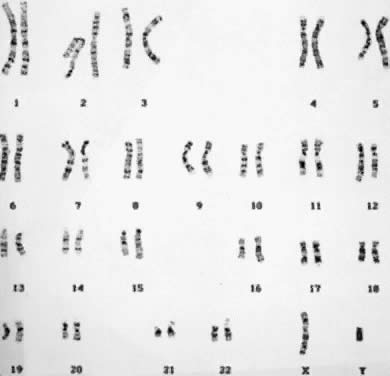
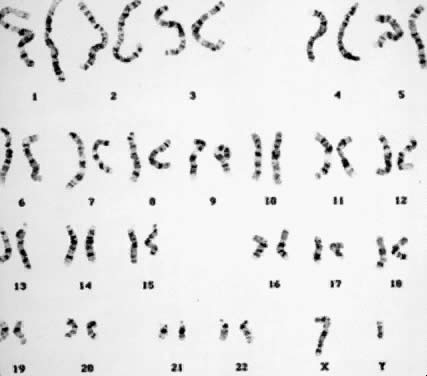
than two times, and the total number of chromosomes in each cell is 47 (Fig. 5). There are several autosomal trisomies (e.g. trisomy 1, trisomy 19) that have not been observed in products of conception
or in liveborns, except possibly with a low percentage of mosaicism. There
are some common trisomic conditions in abortuses that have
never been observed in liveborn infants. For example, trisomy 16 is the
most common trisomy among spontaneous abortions, but it is incompatible
with life. Autosomal trisomies as a group comprise 15% of all chromosomally
abnormal, first-trimester spontaneous abortions. In the case
of trisomy 21 (i.e. Down syndrome), the most common trisomy in liveborns, only one in four
such conceptions reaches term. Not surprisingly, most trisomic conceptions
do not complete term, presumably because of impaired placental function
or fetal maldevelopment. The incidence of numeric chromosome aberrations
in newborns is 1 in 200, or 0.5%. Other less frequent, but
well defined autosomal chromosome syndromes are trisomies 13 and 18. Other
trisomies for chromosomes 8, 9, and 22, also producing definable
syndromes, generally occur in the mosaic state. The congenital malformations
associated with each of these trisomies have been summarized in
several compendiums.29,30,31 Chromosome syndromes caused by trisomy share several cardinal features. Autosomal
syndromes are characterized by IUGR and postnatal growth retardation, a
pattern of dysmorphic features, malformations of several
organ systems, and impaired mental development. Congenital malformations
consistently associated with trisomy are abnormal facies; low-set, malformed
helices; digital anomalies, such as clinodactyly and single
palmar creases; and anomalies of the skeletal, cardiac, and genitourinary
systems. Neonatal survival is compromised by the presence of these
congenital anomalies and shortened life expectancy is characteristic
of trisomies 13, 18, and 21. In the latter case, for example, about one
third of affected infants die during the first year of life, one half
die by age 3 or 4, and the remainder have a reduced life expectancy.32 Females with three X chromosomes (i.e. triplo-X) have clinical features varying from normal to mildly dysmorphic, with
and without infertility, to severe retardation.33 Females with more than three X chromosome are uniformly delayed developmentally, with
variability in physical features ranging from normal to
skeletal and dysmorphic abnormalities, such as microcephaly, radioulnar
synostosis, prognathism, and webbed neck.34 XXY males (i.e. Klinefelter syndrome) are characterized by small, firm testes and seminiferous
tubule dysgenesis, small penis, gynecomastia, incomplete virilization, variable
eunuchoidism, and a tendency to mild mental retardation. In
approximately two thirds of the cases of XXY, the two X chromosomes
are maternally derived and associated with increased maternal age; when
one of the X chromosomes is paternal in origin, maternal age
is not increased.35 In individuals with more complex sex chromosome aneuploidy (e.g. 48,XXXY; 49,XXXXY; 49,XXXYY), multiple somatic anomalies and severe developmental
retardation are common.35 The XYY chromosome syndrome has drawn great interest to the question of
genetic predisposition to criminal behavior since the report of increased
prevalence of the YY constitution among tall and mentally retarded
males in prisons.36,37,38 Studies during the past 25 years have shown that most YY males are phenotypically
normal. Although no anomalous disorders have been observed
in YY males originally identified by random newborn screening, a number
of physical and personality features have been repeatedly described: excessive
height; nodulocystic acne; large deciduous and permanent teeth; subtle
neurologic abnormalities, such as intention tremor and incoordination; genital
abnormalities and possible increased risk for infertility; increased
risk for difficulties with language development; and
a questionably increased risk for behavioral disability.39 Prospective studies of YY children have shown no differences in behavioral
problems compared with XXY children and controls, but the prevalence
of YY males in psychiatric and prison facilities is increased 20 times
above that of the incidence of YY males in the newborn population. Early
reports characterizing YY males as overly aggressive toward other
people have not been confirmed. Reduced intelligence of YY men may
increase their chances of being arrested and incarcerated after any antisocial
act compared with similar crimes committed by XY men. Because
the presence of two Y chromosomes does not impair viability and occurs
with an incidence of 1 in 1000 births, the prenatal diagnosis of an
XYY fetus is not uncommon, and nondirective genetic counseling is indicated.39 Aneuploidy is an integral part of the biologic processes associated with
cancer, particularly those associated with hematologic malignancies, such
as leukemia.40 Cytogenetic analyses can contribute to classifying the type of cancer, monitoring
the progression of the disease, and determining the effects
of treatment. For example, aneuploidy is frequently associated with
blast crisis in chronic myelogenous leukemia. Structural Chromosome Aberrations Structural changes in the normal banding patterns of chromosomes occur
as the result of one of three biologic processes. The most common cause
of structural chromosome aberrations is physical breakage involving
one or more chromosomes. Two other less common causes of structural chromosome
aberrations are unequal crossing over due to mispairing of homologous
chromosomes during meiosis and defective DNA synthesis during
chromosome replication. When a chromosome breaks, the broken ends are sticky or unstable, and cellular repair mechanisms usually rejoin the two ends
promptly (i.e. restitutional healing). In such cases, the occurrence of chromosome breakage
would not be detected. However, if more than one chromosome break
occurs, the repair mechanisms cannot differentiate between sticky ends
and the wrong ends may join (i.e. nonrestitutional healing). Chromosome breakage occurs spontaneously or
is induced by exposure to ionizing radiation or mutagenic chemicals. The
rate of spontaneous, nonrestitutional chromosome breakage exceeds 1 in 1000 gametes, which
is 100 times greater than the spontaneous mutation
rate for single gene loci.1 Understanding the nature and behavior of structural chromosome aberrations
is of considerable clinical importance. Prospective parents carrying
structurally rearranged chromosomes are at significant reproductive
risk for genetically unbalanced conceptions. Structural chromosome aberrations
also have been the primary means for developing a physical map
of the human genome and the first step in the eventual cloning of disease-causing
genes. There are five distinct classes of structural chromosome aberrations. They
are translocation, deletion, duplication, inversion, and isochromosome. TRANSLOCATION. Translocations are interchromosomal exchanges brought about by breakage
and transfer of segments of chromosomes to different locations. There
are three basic types of translocations: reciprocal; centric fusion or Robertsonian; and insertional. In a reciprocal translocation, segments distal to breaks in two chromosomes
are exchanged. The exchange may involve the long or the short arms
of any pair of chromosomes, autosomal or sex (or both), homologous
or nonhomologous. Such an exchange usually does not result in any loss
of genetic material, and the individual is clinically normal and called
a balanced carrier. Nevertheless, a balanced carrier is at significant reproductive risk for
producing chromosomally unbalanced gametes that adversely affect conception
and embryonic development. Figure 9 shows a reciprocal exchange between the long arm of chromosome 4 and the
long arm of chromosome 13 occurring in a balanced carrier. During gametogenesis, homologous
segments of chromosomes normally synapse (i.e. pair together), and in the case of reciprocal translocation between chromosome 4 and 13, this
would produce an association of four chromosomes
in the form of a cross-shaped quadrivalent. At anaphase of meiosis I, depending on the type of chromosome segregation, 12 different
gametes involving chromosomes 4 and 13 are possible, six
from 2:2 segregation (two chromosomes into each daughter cell), and
six from 3:1 segregation. With the former combination, three types
of segregation are possible: alternate, producing one chromosomally normal
gamete and the other that is a chromosomally balanced gamete; adjacent
I, resulting in gametes that carry duplications and deficiencies
for segments of chromosomes 4 and 13; and adjacent II, also producing
gametes with deficiencies and duplications of segments of chromosomes 4 and 13. With 3:1 segregation, six other chromosome combinations are
possible, but the genetic imbalance causes early pregnancy failure, and
in most cases, the pregnancy goes unrecognized. Because centromeres
from homologous chromosomes regularly segregate from one another during
meiosis I of gametogenesis, 2:2 segregation is more likely, particularly
alternate and adjacent I forms. Although in theory gametes with
twelve different chromosome complements are possible, in practice the
actual number is limited by the pattern of chromosome segregation and
their effect on embryonic viability. The actual risk of unbalanced offspring
is always lower than the theoretical risk (Table 2). TABLE 2. Minimal Risk of Chromosomally Unbalanced Conception Detected by
Prenatal Diagnosis for Carriers of Balanced Structural Rearrangements
| | Minimal Risk of |
| | Chromosomally |
| | Unbalanced |
Rearrangement | Carrier | Conception (%) |
Centric fusion 13;14 | Father | 1 |
Centric fusion 13;14 | Mother | 1 |
Centric fusion 14;21 | Father | 1 |
Centric fusion 14;21 | Mother | 11 |
Centric fusion 21;22 | Father | 5 |
Centric fusion 21;22 | Mother | 10 |
Centric fusion 21;21 | Father | 100 |
Centric fusion 21;21 | Mother | 100 |
Reciprocal translocation | Father | 12 |
Reciprocal translocation | Mother | 12 |
Pericentric inversion* | Father | 4 |
Pericentric inversion* | Mother | 8 |
* Does not include the common chromosome 9 pericentric inversion
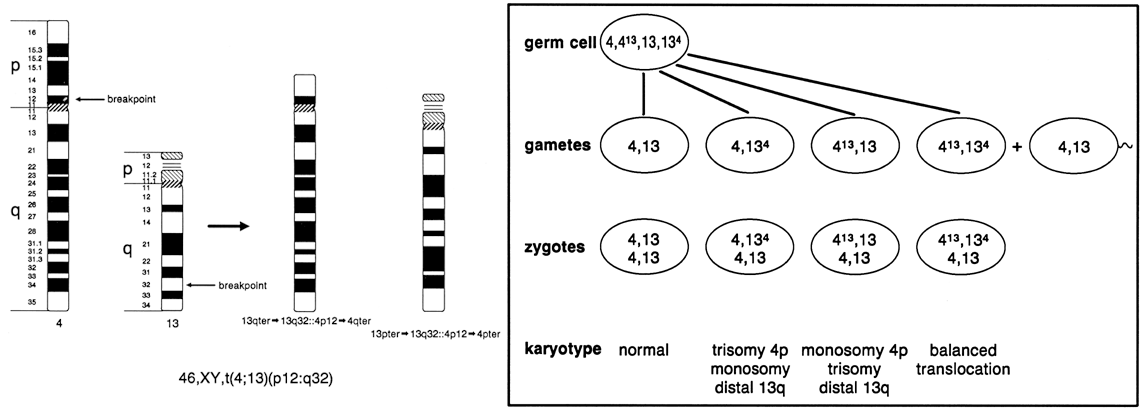 Fig. 9. Meiotic segregation (2:2) in a reciprocal translocation involving chromosomes 4 and 13. Fig. 9. Meiotic segregation (2:2) in a reciprocal translocation involving chromosomes 4 and 13.
|
A de novo, apparently balanced translocation detected in the course of prenatal genetic
diagnostic studies cannot ensure a normal phenotypic outcome. Balanced
translocations have been observed in liveborn infants with congenital
malformations and developmental disabilities whose parental chromosomes
are normal. A survey of centers performing prenatal diagnosis
showed that the risk of an adverse pregnancy outcome with a de novo translocation detected by second-trimester amniocentesis was high, exceeding 9%.41 Although the specific risk of mental retardation and birth defects cannot
be predicted with certainty, empirical data indicate that 1 of 10 sporadic, apparently
balanced translocations may be associated with loss
of normal gene function and abnormal embryonic development. The recurrence
risk (i.e. the chance of a second conception with the same chromosome rearrangement) is
low, although rare instances of gonadal mosaicism for structural
aberrations have been observed. Prenatal diagnostic studies in subsequent
pregnancies are warranted in cases of de novo translocations, despite the low risk of recurrence. Centric fusion, or a Robertsonian translocation, results as a consequence
of chromosome breakage at or near the centromeres of two acrocentric
chromosomes (13, 14, 15, 21, and 22). The breaks usually occur just
above the centromeres, resulting in a single metacentric chromosome with
two centromeres (i.e. dicentric) and a satellite fragment with no centromere (i.e. acentric). Any acentric fragment invariably is lost during cell division. Alternatively, Robertsonian translocations may arise in some cases
as a consequence of crossing over between homologous segments of two
acrocentric chromosomes during meiosis I. In either case, the formation
of a Robertsonian translocation is associated with reduction of chromosome
number by one, with the diploid number being 2N = 45. The genetic
material contained on the short arms of acrocentric chromosomes appears
to have no clinical effects. Despite their loss, individuals who
carry de novo Robertsonian translocations are phenotypically normal. There is, however, an
increased reproductive risk of forming unbalanced gametes during
gametogenesis. Centric fusion of chromosomes 13 and 14 is the most common structural chromosome
aberration in humans, occurring with a frequency of 1 in 1000 individuals.1 The next most common Robertsonian translocation involves chromosome 21 with
chromosomes 13, 14, 15, 21, and 22. As shown in Figure 10, a balanced carrier of a 14;21 centric fusion translocation has 45 chromosomes
and, although healthy, is at significant reproductive risk. When
homologous chromosome segments synapse during meiosis, a trivalent
is formed of the two normal chromosomes, 14 and 21, with the 14;21 translocation
chromosome. At anaphase I, the three chromosomes can segregate
in three ways, producing gametes of six different constitutions. Alternate
segregation is associated with one chromosomally normal gamete
and one balanced gamete. Adjacent I segregation results in one gamete
carrying a duplicated segment of chromosome 14 and one deficient in
the same segment; because both are unbalanced, embryonic and/or fetal
viability are likely to be compromised, and spontaneous abortion is likely. Adjacent
II segregation produces one gamete with duplication of
the long arm of chromosome 21 and one gamete with complete deficiency
of chromosome 21. Although liveborn infants with only one 21 chromosome (from
the other parent) has not been observed, the gamete with duplication
of the long arm of 21 results in a conception with Down syndrome. Although
six different gametic combinations are theoretically possible, only
three types of conceptions are observed: conceptions with a
normal complement of 46 chromosomes; a balanced 14;21 translocation carrier
with 45 chromosomes and a normal pregnancy outcome expected; and
an unbalanced chromosome constitution because of the presence of the 14;21 translocation
and two chromosomes 21, resulting in Down syndrome. These
three types of conceptions do not occur with equal frequency
in the liveborn population, partly because of selection factors favoring
the gametes carrying the 14;21 translocation and partly because of
the sex of the carrier parent. Empiric data obtained in familial forms
of 14;21 translocations indicate that approximately 20% of pregnancies
end in spontaneous abortion; as many as 50% of all liveborn infants
are translocation carriers like one of the parents. If the maternal parent
is the carrier, the risk of a Down syndrome liveborn infant is 11%, whereas
if the paternal parent is the carrier, the risk of Down syndrome
is less than 5%42 (Table 2). 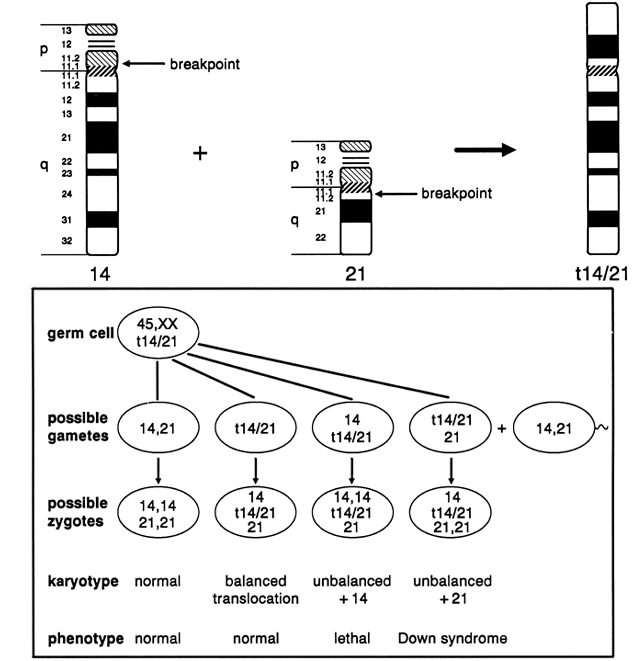 Fig. 10. Meiotic segregation of Robertsonian (centric fusion) translocation involving
chromosomes 14 and 21. Fig. 10. Meiotic segregation of Robertsonian (centric fusion) translocation involving
chromosomes 14 and 21.
|
In approximately 4.5% of all newborn cases, Down syndrome occurs as a consequence
of an unbalanced Robertsonian translocation involving chromosome 21. Chromosome
studies in such newborns, however, show that only
one half of the cases are familial. The remaining cases arise de novo in the meiotic cell division producing the gamete that participated in
fertilization. Down syndrome as a consequence of a Robertsonian 21;21 translocation
presents a unique situation. In approximately 95% of cases, the 21;21 translocation observed in a newborn with Down syndrome
has arisen de novo. The remaining cases most likely are caused by gonadal mosaicism, because
the origin of a balanced carrier for a 21;21 Robertsonian translocation
would require two biologic errors (formation of the translocation
in one parental gamete and loss of a normal chromosome 21 in the other
parental gamete). If centric fusion of the two 21 chromosomes occurred
during gametogenesis, the resulting conception could not be balanced
but rather would be affected with Down syndrome because of the normal
chromosome contributed by the other parent's gamete. If centric
fusion of the two 21 chromosomes occurred postzygotically, presumably
there would be cells with 46 chromosomes and cells with 45 chromosomes
carrying the translocation, with one unlikely exception: formation occurring
before the first cell division of the zygote. What is unique to carriers of 21;21 translocations is that all of their
gametes result in chromosomally unbalanced conceptions (Fig. 11). Approximately one half of the conceptions are affected with Down syndrome, based
on the contribution of the 21;21 translocation from one parent
and a normal 21 chromosome from the other parent. The chromosome
constitution of the remaining conceptions has only one chromosome 21 (monosomy 21), and
will not be viable. If all of the cells of a potential
parent carry the 21;21 translocation in a balanced state, prenatal
diagnosis would be of no value, because all pregnancies are chromosomally
unbalanced and developmentally compromised. When Down syndrome occurred
secondary to a Robertsonian 21;21 translocation and parental karyotypes
were normal, there were no recurrences of Down syndrome.43 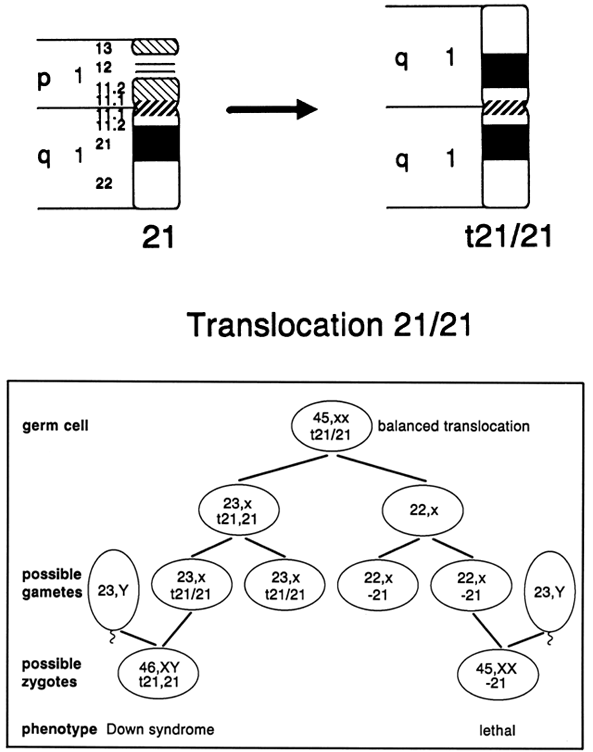 Fig. 11. Meiotic segregation of t(21;21) in a balanced translocation carrier. Fig. 11. Meiotic segregation of t(21;21) in a balanced translocation carrier.
|
DELETION. A deletion is the loss of any part of a chromosome. It can occur in several
ways. A terminal deletion occurs when a single break in a chromosome
produces centric and acentric chromosomes. The latter chromosome, lacking
a centromere, is lost in the next cell generation, whereas when
the centric chromosome duplicates, the broken ends of the chromatids
can fuse together, forming a dicentric chromosome. More commonly, an
interstitial deletion arises from the breakage at two points along a
chromosome (Fig. 12A). If the chromosome segment between the two sites of breakage does not
include the centromere, the acentric fragment so formed will be lost. If
breakage occurs in both chromosome arms, the terminal, acentric ends
are lost, and the two proximal ends may unite to form a ring chromosome. A
ring chromosome with a centromere is usually able to complete
mitosis successfully, although if sister-strand crossing over occurs, a
ring chromosome that is twice the size of the original ring chromosome, with
two centromeres, is formed. Deletion of segments of chromosomes
can occur as a consequence of adjacent I and II disjunction involving
a parental translocation. 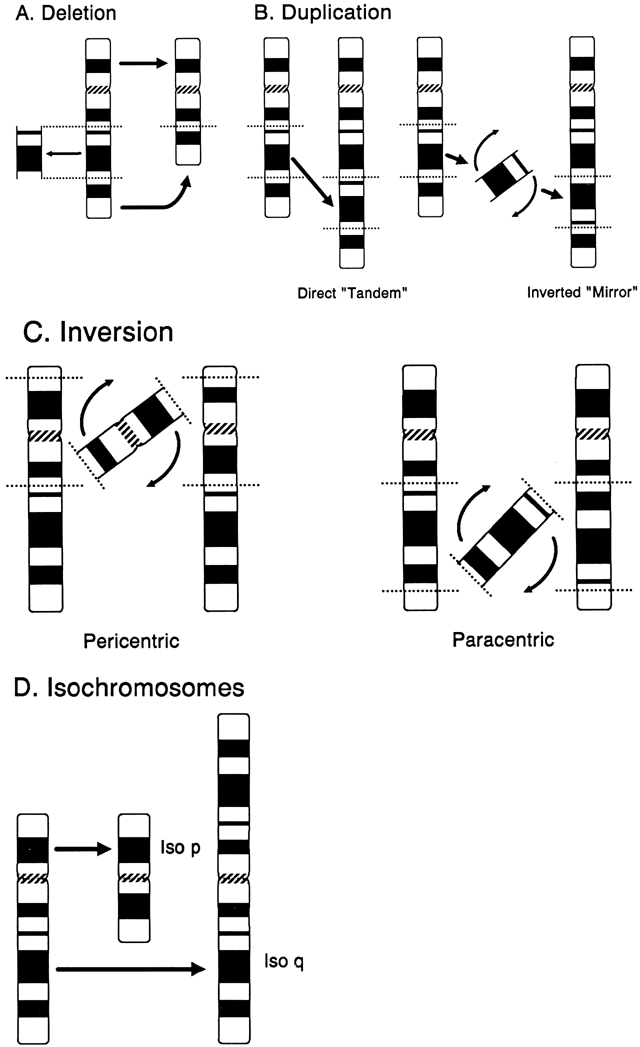 Fig. 12. Structural chromosome abnormalities. A. Deletion. B. Duplications: direct (tandem) and inverted (mirror). C. Inversions: pericentric and paracentric. D. Isochromosomes. Fig. 12. Structural chromosome abnormalities. A. Deletion. B. Duplications: direct (tandem) and inverted (mirror). C. Inversions: pericentric and paracentric. D. Isochromosomes.
|
The smallest loss of chromosomal material detected microscopically is approximately 400,000 base
pairs.1 Conceptions with visible deletions can be monosomic for a large number
of genes. With autosomal deletions that are viable, developmental disabilities
with mental retardation are to be expected. Specific syndromes
associated with deletions include the cri-du-chat syndrome with visible
loss of genetic material from the short arm of chromosome 5; the
Wolf-Hirschhorn syndrome with deletion in the short arm of chromosome 4; and
the Prader-Willi syndrome, with paternally derived deletion of 15q11. Part
of a group of syndromes designated as contiguous gene syndromes44 includes conditions that occasionally have been associated with small
but cytogenetically detectable deletions or duplications. Included in
this group are deletions of 8q24 in Langer-Giedion syndrome,45 deletions of 11p13 associated with aniridia and Wilms tumor,46 deletions of 13q14 associated with retinoblastoma,47 deletions of 17p13 in the Miller-Dieker syndrome,48 and deletions of 22q11 in DiGeorge syndrome.49 When cytogenetic analysis does not show a visible deletion, molecular
analysis can demonstrate the loss of specific gene sequences representing
submicroscopic deletions of the band or bands previously associated
with the specific syndrome. The ability to apply molecular techniques
for the cloning of the gene responsible for Duchenne and Becker muscular
dystrophies, for example, first required determining its physical
location, which was accomplished through deletion mapping.50 DUPLICATION. Duplication of any segment of a chromosome may arise as the result of
unequal crossing over during meiosis or from meiotic events in a parent
with a translocation, inversion, or isochromosome (Fig. 12B). In general, duplications as a form of chromosomal imbalance appear to
be better tolerated clinically than deletions. At the molecular level, duplications
in the form of repeats of specific nucleotide sequences
appear to play a primary role in such gene disorders as the fragile-X
syndrome, myotonic dystrophy, and the Kennedy syndrome and may be an
important feature in many other human genes.51 For example, the fragile-X syndrome is caused by a heritable unstable
DNA sequence because of changes in copy number of a trinucleotide repeat, CGG.52 The overall incidence of visible deletions, duplications, or a combination
of the two in newborns is estimated at 1 case 1 per 2000.53 For any chromosomal duplication or deletion detected prenatally or postnatally, parental
chromosomes must be examined to exclude a balanced
structural rearrangement. If parental karyotypes are normal, the recurrence
risk is not increased above the general population incidence, with
the rare exception of gonadal mosaicism. INVERSION. Inversions arise from two chromosome breaks occurring in the same chromosome, followed
by a rotation of 180 degrees of the segment between the
breaks (Fig. 12C). If both breaks occur in the same arm so that the centromere is not included, the
inversion is called paracentric. If the breaks are on either side of the centromere, the inversion is called pericentric. The change in gene order has not been associated with any clinical abnormalities, but
inversions place their carriers at reproductive risk as
a consequence of their structure and behavior during meiosis. Inversions
may interfere with the pairing of homologous chromosomes and thereby
reduce crossing over within the inverted segment, but for synapsis
to occur, one member must form a loop in the region of the inversion (Fig. 13A). If crossing over occurs within the loop of a paracentric inversion, a
dicentric chromatid and an acentric fragment are formed, both of which
are unstable and unlikely to result in abnormal offspring. If a single
crossover occurs within the loop of a pericentric inversion, the resulting
two chromatids have complementing deletions and duplications, and
abnormal offspring may result (Fig. 13B). 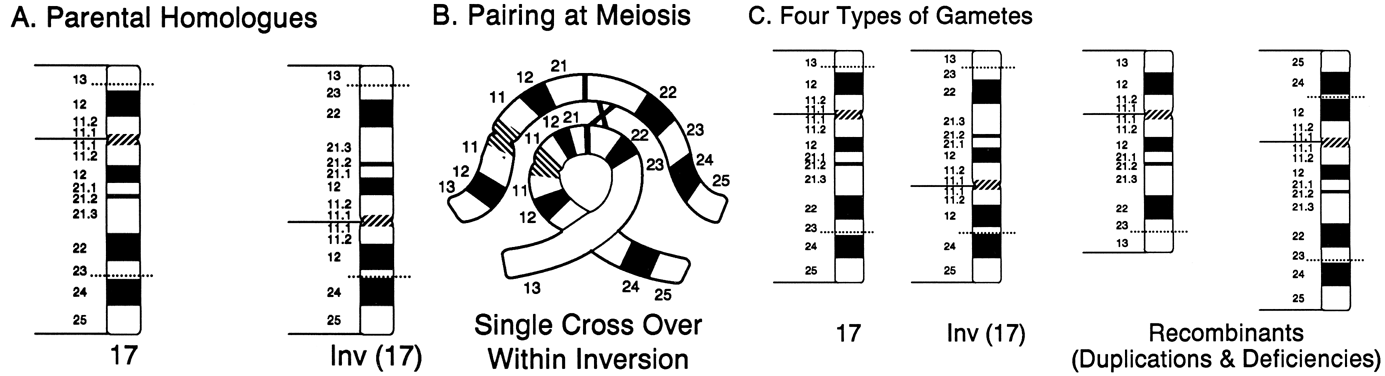 Fig. 13. Products of meiotic crossing over in a pericentric inversion. A. Parental homologues. B. Loop formation in meiotic pairing of normal and inverted chromosomes. Only
two of the four chromatids are shown. C. Exchange products, both with duplications and deletions. Fig. 13. Products of meiotic crossing over in a pericentric inversion. A. Parental homologues. B. Loop formation in meiotic pairing of normal and inverted chromosomes. Only
two of the four chromatids are shown. C. Exchange products, both with duplications and deletions.
|
The closer both breakpoints are to the ends of the chromosomes, the greater
is the chance that the pregnancy will come to term. Paracentric inversions
are not an indication for prenatal diagnosis, because if crossing
over does occur within the inversion loop, the degree of genetic
imbalance is incompatible with viability in all but the most exceptional
instances. Carriers of pericentric inversions, however, are at risk
for viable, chromosomally unbalanced offspring, especially when a large
inversion is involved. Empirically, this risk is 8% for maternal carriers
and 4% for paternal carriers53 (Table 2). One exception is worthy of mention: small pericentric inversions of
chromosome 9. These common inversions, which are present in 1% of the
general population, have not been associated with abnormal offspring as
a consequence of crossing over within the inverted segment. ISOCHROMOSOME. An isochromosome is a structurally altered chromosome in which there is
deletion of one entire arm and complete duplication of the other arm (Fig. 12D). This type of chromosomal aberration most commonly arises by misdivision
of the centromere in the transverse direction. Another mechanism involves
the simultaneous breakage of both chromatids (i.e. isochromatid breakage), followed by fusion of the centromeric portions
and resulting in a dicentric chromosome. When the isochromosome is dicentric, one
centromere apparently becomes nonfunctional so that the isochromosome
routinely segregates normally during cell division. Isochromosomes
for the long arms of the X and Y chromosomes have been observed
in liveborn infants, whereas for other chromosomes, an isochromosome
usually leads to an early spontaneous abortion, with two rare exceptions—isochromosomes
for the short arms of chromosomes 9 and 12.1 Special Characteristics of the X and Y Chromosomes The X and Y chromosomes exhibit certain unique biologic properties that
are clinically relevant to the practice of obstetrics and gynecology. X INACTIVATION. According to the Lyon hypothesis, in somatic cells, one of the two X chromosomes
becomes inactivated early in embryonic life. The inactivation
is random; the maternal or the paternal X chromosome may be inactivated. X
inactivation is virtually complete (i.e. almost all of the genes on the X chromosome are inactivated). X chromosome
inactivation is permanent and clonally propagated, and if a maternally
derived X chromosome is inactivated in a parental cell, an active
paternally derived X chromosome is expressed in all of the daughter
cells, whereas the maternally derived X remains inactive.54 The Lyon hypothesis explains the phenomenon of dosage compensation in
females wherein the amount of gene product of X-coded genes is the same
in females with two X chromosomes as in males with one X chromosome. Several important cytogenetic observations contributed to the development
of the Lyon hypothesis. First and foremost was cytologic evidence of
the existence of sexual dimorphism: only the nuclei of female cells
contain a dark-staining chromatin body approximately 1 μm in diameter
in juxtaposition to the nuclear membrane (the X-chromatin mass, or
Barr body). Subsequent studies in humans with more than two X chromosomes
showed that the number of X-chromatin bodies was equal to the number
of X chromosomes minus 1. Whereas 46,XY males exhibit no X-chromatin
mass in their cells during interphase and 46,XX females exhibit one
X-chromatin mass, 47,XXX and 48,XXXY individuals have two X-chromatin
bodies in their cells. Studies also showed that during prophase, one of
the two X chromosomes appeared to be heteropyknotic (i.e. dense and dark-staining), which suggests gene inactivity. In vitro studies monitoring the incorporation of bromodeoxyuridine (BUdr) into
DNA indicated differential replication of the two X chromosomes: the inactive
X chromosome was the last chromosome to replicate its DNA during
the S (i.e. synthesis) phase of mitosis and hence the term, “late replicating, was applied to the inactive X chromosome.55 In a patient with 49,XXXXX aneuploidy, the cells exhibit the presence
of four X-chromatin bodies, four heteropyknotic X chromosomes during prophase, and
four late-replicating X chromosomes in the S phase of mitosis, all
of which provide cytologic evidence of genetically inactive
X chromosomes. The Lyon hypothesis holds the key to several medically significant problems. It
provides the biologic basis for the greater variation in clinical
manifestations of X-linked genes and diseases in heterozygous females, compared
with hemizygous males. In the same manner, it explains
past difficulties in using biochemical techniques to identify carrier
females of X-linked gene mutations and why this approach has been replaced, wherever
possible, by X-linked DNA markers. Lyonization has provided
insight into sex determination in humans because the gene action
of the Y chromosome is basically interacting with only one functional
X chromosome, despite the presence of two or more X chromosomes. A paradox remains, however, in that individuals with more than two X chromosomes
may not be developmentally normal, and the more X chromosomes
a patient has, the greater the likelihood of mental retardation. X chromosome
inactivation during human embryonic development is believed
to occur during the blastocyst stage of implantation. An interesting observation
whose biologic significance has yet to be determined is the
fact that the paternal X chromosome is inactivated preferentially in
placental tissue and other extraembryonic membranes. X chromosome inactivation
also has been used as a marker in studies of differentiation
and malignancy. For example, in a study of uterine fibroids in women who
were heterozygous for glucose-6-phosphate dehydrogenase alleles, A
and B, each fibroid tumor expressed only the A or B type, but never both. This
finding indicated that each tumor arose from the clonal propagation
of a single cell rather than a multicentric origin.53 The existence of a specific site on the X chromosome responsible for the
process of X chromosome inactivation was first inferred from studies
of X autosome translocations in humans and from mapping of a murine locus, Xce, which influences preferential inactivation of an X chromosome in a cis-acting manner.56 The location of the X inactivation center (XIC) in humans has been determined to be located at Xq13, although the DNA
sequence itself remains unresolved. A transcript that is exclusively transcribed
from the inactive X chromosome has been identified, the X-inactive-specific
transcript (XIST).57 A close relation exists between inactivation of the X chromosome and expression
of XIST.58,59,60 XIST is expressed during the period in male germ cell development in which
the single X chromosome is transiently inactivated and XIST expression
ceases at a time in female germ cell development when the inactive
X chromosome becomes reactivated. A number of questions remain.61 Does XIST expression lead to inactivation of the X chromosome, or does
XIST expression occur because the X chromosome has become inactivated? How
does the cell maintain one and only one active X chromosome? If
XIST has a functional role in X inactivation, how is this accomplished? Answers
to these questions will further understanding of the nature
of X chromosome disorders and possibly lead to methods of amelioration. Y CHROMOSOME. In humans, the Y chromosome is responsible for normal male development. Phenotypic-karyotypic
correlations in males with structural aberrations
of the Y chromosome first contributed to localizing genes affecting
stature and testicular development. The entire Y chromosome has been
cloned using molecular techniques,62 setting the stage for the complete gene mapping of the Y chromosome.63 The presence of an XY karyotype or a structurally modified Y chromosome
in a patient with a female habitus can present difficult clinical challenges. In
the case of 45,X/46,XY chromosome mosaicism, the phenotype
ranges from almost normal males with cryptorchidism or penile hypospadias
to females indistinguishable from those with 45,X (i.e. gonadal dysgenesis). A diagnosis of 45,X/46,XY is likely if the patient
has a unilateral streak gonad and a contralateral testis, or bilateral
dysgenetic testes and mullerian derivatives.64 Approximately 5% of patients with gonadal dysgenesis and unambiguous external
genitalia have cells containing a Y chromosome in addition to
cells with 45,X. The dysgenetic gonads may undergo transformation into
a gonadoblastoma or dysgerminoma. Most 45,X/46,XY persons have ambiguous
genitalia and gonads consisting of a unilateral streak one side and
a contralateral dysgenetic testis (i.e. mixed gonadal dysgenesis). The risk of neoplastic transformation ranges
between 15% and 20%. Breast enlargement in a 45,X/46,XY individual with
ambiguous external genitalia is clinical evidence of a gonadoblastoma
or dysgerminoma.64 A paradox exists in that, when 45,X/46,XX is detected prenatally, the
fetus invariably has normal male external genitalia, with the only abnormalities
being hypospadias or unilateral cryptorchidism.65 Subjects with the XY form of pure gonadal dysgenesis (i.e. Swyer syndrome) have bilateral streak gonads, female external genitalia, müllerian
derivatives (i.e. uterus and fallopian tubes), and no somatic abnormalities.66 The familial form of Swyer syndrome is inherited as an X-linked recessive
or a male-limited autosomal dominant gene. In many sporadic forms
and some familial cases, microdeletions of the Y short arm containing
the testisdetermining factor (TDF) can be responsible for Swyer syndrome.67 In addition to the effects of hypogonadism and infertility, the XY form
of pure gonadal dysgenesis is associated with a 30% lifetime risk of
gonadal neoplasia. Fragile Sites Fragile sites describe the cytologic observation that segments of chromosomal
material appear as nonstaining gaps of variable width, usually
involving both chromatids. A fragile site is always observed at the same
point on the chromosome in cells studied from a subject. It is inherited
in a mendelian codominant fashion. Molecular studies are beginning
to elucidate the structural and functional natures of fragile sites. A
fragile site may represent a specific segment of a chromosome that
does not undergo normal condensation in mitosis. The loci of several
oncogenes and the sites of chromosomal breakage that lead to structural
rearrangements correlate with sites of fragility. The most clinically
relevant fragile site is that of the X chromosome at band q27, which
accounts for one third of all mentally retarded males and females (Fig. 14). The effect on the human population is significant, because the frequency
of the fragile-X syndrome is approximately 1 in 1500. 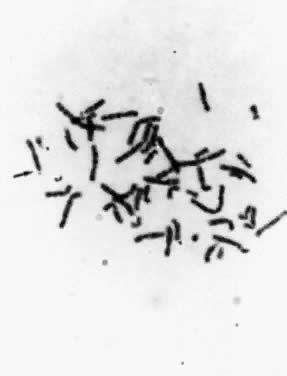 Fig. 14. Fragile-X chromosome. Metaphase spread from the peripheral blood lymphocyte
culture of a male subject with fragile-X syndrome. Fig. 14. Fragile-X chromosome. Metaphase spread from the peripheral blood lymphocyte
culture of a male subject with fragile-X syndrome.
|
Molecular studies have resulted in identification of a gene, FMR1, containing a trinucleotide repeat sequence, CGG, coincident with the fragile-X
syndrome.52,68 In fragile-X syndrome, a CGG repeat in the 5' untranslated region
of the FMR-1 transcript normally is polymorphic, displaying alleles
ranging from 6 to 60 repeats (mean, 29). This repeat is unstable in fragile-X
families with repeat lengths varying from 60 to approximately 200 triplets
in nonpenetrant carrier females and massively expanded well
beyond 200 repeats, often up to 1000 triplets, in penetrant males. About
one half of carrier females with more than 200 copies are mentally
impaired. Expansion beyond 200 repeats is accompanied by methylation
of the repeat region, which results in transcriptional silencing of
the gene and the clinical expression of the fragile-X syndrome. Increases
in the length of the CGG repeat in fragile-X carrier females is correlated
with proportional increases in risk to full expansions and penetrant
offspring.69 This phenomenon is similar to genetic anticipation, in which increasing clinical involvement and decreasing age of onset
occur in successive generations. |










 33 years) undergoing first-trimester chorionic villus sampling (CVS), the
overall frequency of chromosome aberrations was 2.5%. The procedure
was limited to women with viable pregnancies whose growth parameters
were compatible with gestational age. For the same women undergoing second-trimester
amniocentesis, 1.5% had karyotypically abnormal pregnancies.
33 years) undergoing first-trimester chorionic villus sampling (CVS), the
overall frequency of chromosome aberrations was 2.5%. The procedure
was limited to women with viable pregnancies whose growth parameters
were compatible with gestational age. For the same women undergoing second-trimester
amniocentesis, 1.5% had karyotypically abnormal pregnancies.